Standardization description
The in-house standardization uses
the Indigo toolkit.
It consists in the following operations:
- Discarding compounds if the number of atoms is > maxlength
- Normalizing structure using Indigo: neutralizes charges, resolves 5-valence Nitrogen, removes hydrogens
- Dearomatizing: converts molecules/reactions to Kekule form
- Standardizing with the following options:
- Keeping only the largest fragment in the molecule.
- Removing fragments that consist of only a single heavy atom.
- Setting all atoms and bonds to NoStereo.
- Removing all relative stereo groupings.
- Setting all atoms and bonds marked UnknownStereo to NoStereo.
- Setting all atoms marked UnknownStereo to NoStereo.
- Setting all bonds marked UnknownStereo to NoStereo.
- Clearing any atom valence query features and resets all implicit hydrogen counts to their standard values.
- Setting the charges on a molecule to a standard form.
- Applying the following 3 reactions, added in-house :
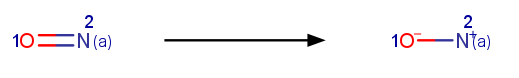

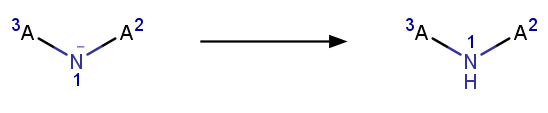
- Aromatizing: Converts molecules/reactions back to aromatic form
- Computing the canonical SMILES (also known as absolute SMILES) string .